The Pathogen Genomics Symposium, hosted by the Robert J. Havey, MD Institute for Global Health’s Center for Pathogen Genomics and Microbial Evolution, is an annual event that includes a day of scholarly events, including research talks, flash talks, keynote address, poster session, and reception. The 2026 event will take place on Monday, May 4th in Lurie Medical Research Center (303 E Superior St, Chicago, IL 60611.)
Center for Pathogen Genomics and Microbial Evolution
Providing specialized expertise in pathogen-specific sequence analysis and bioinformatics for emerging and ongoing infectious disease threats.
The development of next generation DNA sequencing approaches has revolutionized genomics. As these technologies have become more affordable, easier to use and widely available, there has been an explosion of genomic sequence generated with hundreds of thousands of bacterial, viral and fungal genome sequences currently available through public databases. The wealth of microbial sequence data available, however, has far outpaced expertise and tools for analysis of these data. Owing in part to the considerable biological diversity among microbes - including bacteria, fungi and viruses - microbial genome sequence analysis has been difficult to standardize. Often, analysis approaches are not “one-size-fits-all” and a technique that is successful for studying one organism may not be directly applicable to another. To fully leverage the power of these tools, diverse expertise is required across several fields including microbiology, infectious disease clinical care, epidemiology and computational bioinformatics.
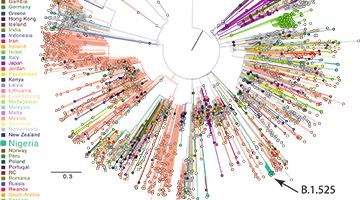
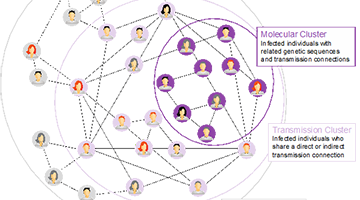
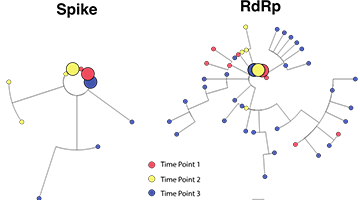
When properly harnessed through an integrated pipeline from sample acquisition to sequencing and bioinformatic analysis, pathogen genomics and molecular epidemiology can define the determinants of pathogen transmission, pathogenesis and evolution and so inform best practices in clinical care and public health. Indeed, the essentiality of rapid and effective microbial sequence analysis has been underscored by the global COVID-19 pandemic. Novel and creative approaches to SARS-CoV-2 viral sequencing and analysis have been central to important discoveries throughout the pandemic that have dictated diagnostic paradigms, facilitated drug discovery, charted routes of transmission and helped define best public health practices.
Northwestern has unmatched abilities in this field that is critical to the national and global efforts to track and study infectious disease threats. Implementing and expanding microbial genomic sequencing and innovative analysis capabilities pays dividends in infections prevented and lives saved.”
–Egon A. Ozer, MD, PhD, Director, Center for Pathogen Genomics and Microbial Evolution

Our Mission and Vision
Why It's Important
Microbial genomics: the study of the complete genetic code of pathogenic organisms
Pathogen Genomics Symposium
Center Leadership
Services
The Lenny Family Global Health Fund In Honor of Robert J. Havey, MD, and In Memory of Andrew J. McKenna
This funding mechanism, established in 2025, provides support to the Center for Pathogen Genomics and Microbial Evolution to develop international strategies to address the growing global burden of infections caused by antimicrobial resistant bacteria. Infections caused by resistant bacteria are increasingly difficult to treat with conventional antibiotics and are predicted to contribute to more than 8 million global deaths annually by the year 2050. A global approach using state-of-the-art technologies for tracking the emergence and spread of antimicrobial resistant infections around the world is crucial to develop effective strategies for prevention and new therapies for treatment of these dangerous diseases.
Supported by a $1.2M investment, the Global Genomic Surveillance of Antimicrobial Resistance (GENOSAR) project is building capacity to detect and track antimicrobial resistant bacteria in low- and middle-income countries with high burdens of infections. Working with partners at sentinel sites in the countries of Nigeria, Pakistan, Peru, and South Africa, the GENOSAR project establishes local programs for the surveillance of antibiotic-resistant bacteria by developing laboratory infrastructure for microbial genome sequencing and building capacity for the analysis of genome sequence data. The information collected by the sentinel sites will include measures of the genetic diversity of bacteria that cause infections and the mutations they develop to become resistant to antibiotics. Using computational modeling, these data will be analyzed to understand how antibiotic-resistant bacteria develop and spread and how these infections can be more effectively prevented and treated.
Featured Projects

Successful Clinical Response in Pneumonia Therapy (SCRIPT)
Northwestern PIs: Richard Wunderink, MD; Alan Hauser, MD, PhD
This U19 systems biology center funded by the NIAID investigates host/pathogen interactions that result in unsuccessful responses to therapy in severe pneumonia. The study combines clinical phenomics, host transcriptomics and epigenomics, pathogen genomics and alveolar metagenomics to model disease dynamics during ventilator-associated pneumonia.